GPyTorch regression with derivative information¶
Introduction¶
In this notebook, we show how to train a GP regression model in GPyTorch of an unknown function given function value and derivative observations. We consider modeling the function:
using 50 value and derivative observations.
[1]:
import torch
import gpytorch
import math
from matplotlib import pyplot as plt
import numpy as np
%matplotlib inline
%load_ext autoreload
%autoreload 2
Setting up the training data¶
We use 50 uniformly distributed points in the interval \([0, 5 \pi]\)
[2]:
lb, ub = 0.0, 5*math.pi
n = 50
train_x = torch.linspace(lb, ub, n).unsqueeze(-1)
train_y = torch.stack([
torch.sin(2*train_x) + torch.cos(train_x),
-torch.sin(train_x) + 2*torch.cos(2*train_x)
], -1).squeeze(1)
train_y += 0.05 * torch.randn(n, 2)
Setting up the model¶
A GP prior on the function values implies a multi-output GP prior on the function values and the partial derivatives, see 9.4 in http://www.gaussianprocess.org/gpml/chapters/RW9.pdf for more details. This allows using a MultitaskMultivariateNormal and MultitaskGaussianLikelihood to train a GP model from both function values and gradients. The resulting RBF kernel that models the covariance between the values and partial derivatives has been implemented in RBFKernelGrad and the
extension of a constant mean is implemented in ConstantMeanGrad.
The RBFKernelGrad is generally worse conditioned than the RBFKernel, so we place a lower bound on the noise parameter to keep the smallest eigenvalues of the kernel matrix away from zero.
[3]:
class GPModelWithDerivatives(gpytorch.models.ExactGP):
def __init__(self, train_x, train_y, likelihood):
super(GPModelWithDerivatives, self).__init__(train_x, train_y, likelihood)
self.mean_module = gpytorch.means.ConstantMeanGrad()
self.base_kernel = gpytorch.kernels.RBFKernelGrad()
self.covar_module = gpytorch.kernels.ScaleKernel(self.base_kernel)
def forward(self, x):
mean_x = self.mean_module(x)
covar_x = self.covar_module(x)
return gpytorch.distributions.MultitaskMultivariateNormal(mean_x, covar_x)
likelihood = gpytorch.likelihoods.MultitaskGaussianLikelihood(num_tasks=2) # Value + Derivative
model = GPModelWithDerivatives(train_x, train_y, likelihood)
The model training is similar to training a standard GP regression model
[4]:
# this is for running the notebook in our testing framework
import os
smoke_test = ('CI' in os.environ)
training_iter = 2 if smoke_test else 50
# Find optimal model hyperparameters
model.train()
likelihood.train()
# Use the adam optimizer
optimizer = torch.optim.Adam(model.parameters(), lr=0.1) # Includes GaussianLikelihood parameters
# "Loss" for GPs - the marginal log likelihood
mll = gpytorch.mlls.ExactMarginalLogLikelihood(likelihood, model)
for i in range(training_iter):
optimizer.zero_grad()
output = model(train_x)
loss = -mll(output, train_y)
loss.backward()
print('Iter %d/%d - Loss: %.3f lengthscale: %.3f noise: %.3f' % (
i + 1, training_iter, loss.item(),
model.covar_module.base_kernel.lengthscale.item(),
model.likelihood.noise.item()
))
optimizer.step()
Iter 1/50 - Loss: 71.141 lengthscale: 0.693 noise: 0.693
Iter 2/50 - Loss: 69.100 lengthscale: 0.744 noise: 0.644
Iter 3/50 - Loss: 66.347 lengthscale: 0.797 noise: 0.598
Iter 4/50 - Loss: 64.771 lengthscale: 0.845 noise: 0.554
Iter 5/50 - Loss: 63.744 lengthscale: 0.886 noise: 0.513
Iter 6/50 - Loss: 61.682 lengthscale: 0.928 noise: 0.474
Iter 7/50 - Loss: 59.820 lengthscale: 0.961 noise: 0.437
Iter 8/50 - Loss: 57.801 lengthscale: 0.987 noise: 0.402
Iter 9/50 - Loss: 56.894 lengthscale: 1.004 noise: 0.370
Iter 10/50 - Loss: 54.522 lengthscale: 1.010 noise: 0.340
Iter 11/50 - Loss: 53.263 lengthscale: 1.006 noise: 0.311
Iter 12/50 - Loss: 50.900 lengthscale: 0.998 noise: 0.285
Iter 13/50 - Loss: 49.472 lengthscale: 0.986 noise: 0.260
Iter 14/50 - Loss: 47.405 lengthscale: 0.980 noise: 0.238
Iter 15/50 - Loss: 46.851 lengthscale: 0.982 noise: 0.217
Iter 16/50 - Loss: 43.638 lengthscale: 0.991 noise: 0.198
Iter 17/50 - Loss: 42.900 lengthscale: 1.002 noise: 0.180
Iter 18/50 - Loss: 39.969 lengthscale: 1.021 noise: 0.164
Iter 19/50 - Loss: 38.408 lengthscale: 1.040 noise: 0.149
Iter 20/50 - Loss: 35.881 lengthscale: 1.059 noise: 0.135
Iter 21/50 - Loss: 34.669 lengthscale: 1.078 noise: 0.122
Iter 22/50 - Loss: 32.928 lengthscale: 1.097 noise: 0.111
Iter 23/50 - Loss: 30.690 lengthscale: 1.113 noise: 0.100
Iter 24/50 - Loss: 28.567 lengthscale: 1.127 noise: 0.091
Iter 25/50 - Loss: 27.540 lengthscale: 1.138 noise: 0.082
Iter 26/50 - Loss: 24.865 lengthscale: 1.142 noise: 0.074
Iter 27/50 - Loss: 23.273 lengthscale: 1.141 noise: 0.067
Iter 28/50 - Loss: 20.533 lengthscale: 1.147 noise: 0.061
Iter 29/50 - Loss: 19.787 lengthscale: 1.144 noise: 0.055
Iter 30/50 - Loss: 16.676 lengthscale: 1.146 noise: 0.050
Iter 31/50 - Loss: 14.890 lengthscale: 1.151 noise: 0.045
Iter 32/50 - Loss: 13.735 lengthscale: 1.158 noise: 0.040
Iter 33/50 - Loss: 11.772 lengthscale: 1.171 noise: 0.036
Iter 34/50 - Loss: 9.266 lengthscale: 1.182 noise: 0.033
Iter 35/50 - Loss: 7.507 lengthscale: 1.193 noise: 0.030
Iter 36/50 - Loss: 5.724 lengthscale: 1.195 noise: 0.027
Iter 37/50 - Loss: 5.030 lengthscale: 1.195 noise: 0.024
Iter 38/50 - Loss: 1.297 lengthscale: 1.207 noise: 0.022
Iter 39/50 - Loss: -0.072 lengthscale: 1.211 noise: 0.020
Iter 40/50 - Loss: -2.852 lengthscale: 1.208 noise: 0.018
Iter 41/50 - Loss: -4.053 lengthscale: 1.200 noise: 0.016
Iter 42/50 - Loss: -5.160 lengthscale: 1.198 noise: 0.014
Iter 43/50 - Loss: -8.217 lengthscale: 1.208 noise: 0.013
Iter 44/50 - Loss: -8.949 lengthscale: 1.216 noise: 0.012
Iter 45/50 - Loss: -11.805 lengthscale: 1.228 noise: 0.011
Iter 46/50 - Loss: -14.472 lengthscale: 1.230 noise: 0.010
Iter 47/50 - Loss: -17.141 lengthscale: 1.228 noise: 0.009
Iter 48/50 - Loss: -16.575 lengthscale: 1.215 noise: 0.008
Iter 49/50 - Loss: -18.488 lengthscale: 1.204 noise: 0.007
Iter 50/50 - Loss: -20.305 lengthscale: 1.207 noise: 0.007
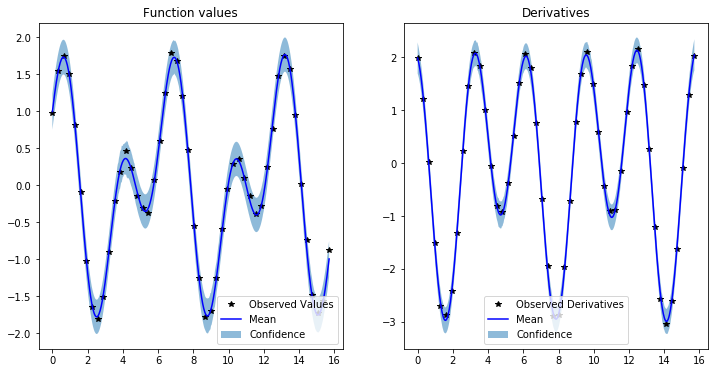
Model predictions are also similar to GP regression with only function values, butwe need more CG iterations to get accurate estimates of the predictive variance
[5]:
# Set into eval mode
model.train()
model.eval()
likelihood.eval()
# Initialize plots
f, (y1_ax, y2_ax) = plt.subplots(1, 2, figsize=(12, 6))
# Make predictions
with torch.no_grad(), gpytorch.settings.max_cg_iterations(50):
test_x = torch.linspace(lb, ub, 500)
predictions = likelihood(model(test_x))
mean = predictions.mean
lower, upper = predictions.confidence_region()
# Plot training data as black stars
y1_ax.plot(train_x.detach().numpy(), train_y[:, 0].detach().numpy(), 'k*')
# Predictive mean as blue line
y1_ax.plot(test_x.numpy(), mean[:, 0].numpy(), 'b')
# Shade in confidence
y1_ax.fill_between(test_x.numpy(), lower[:, 0].numpy(), upper[:, 0].numpy(), alpha=0.5)
y1_ax.legend(['Observed Values', 'Mean', 'Confidence'])
y1_ax.set_title('Function values')
# Plot training data as black stars
y2_ax.plot(train_x.detach().numpy(), train_y[:, 1].detach().numpy(), 'k*')
# Predictive mean as blue line
y2_ax.plot(test_x.numpy(), mean[:, 1].numpy(), 'b')
# Shade in confidence
y2_ax.fill_between(test_x.numpy(), lower[:, 1].numpy(), upper[:, 1].numpy(), alpha=0.5)
y2_ax.legend(['Observed Derivatives', 'Mean', 'Confidence'])
y2_ax.set_title('Derivatives')
None
[ ]:
[ ]: