GPyTorch Regression Tutorial¶
Introduction¶
In this notebook, we demonstrate many of the design features of GPyTorch using the simplest example, training an RBF kernel Gaussian process on a simple function. We’ll be modeling the function
with 100 training examples, and testing on 51 test examples.
Note: this notebook is not necessarily intended to teach the mathematical background of Gaussian processes, but rather how to train a simple one and make predictions in GPyTorch. For a mathematical treatment, Chapter 2 of Gaussian Processes for Machine Learning provides a very thorough introduction to GP regression (this entire text is highly recommended): http://www.gaussianprocess.org/gpml/chapters/RW2.pdf
[7]:
import math
import torch
import gpytorch
from matplotlib import pyplot as plt
%matplotlib inline
%load_ext autoreload
%autoreload 2
The autoreload extension is already loaded. To reload it, use:
%reload_ext autoreload
Set up training data¶
In the next cell, we set up the training data for this example. We’ll be using 100 regularly spaced points on [0,1] which we evaluate the function on and add Gaussian noise to get the training labels.
[8]:
# Training data is 100 points in [0,1] inclusive regularly spaced
train_x = torch.linspace(0, 1, 100)
# True function is sin(2*pi*x) with Gaussian noise
train_y = torch.sin(train_x * (2 * math.pi)) + torch.randn(train_x.size()) * math.sqrt(0.04)
Setting up the model¶
The next cell demonstrates the most critical features of a user-defined Gaussian process model in GPyTorch. Building a GP model in GPyTorch is different in a number of ways.
First in contrast to many existing GP packages, we do not provide full GP models for the user. Rather, we provide the tools necessary to quickly construct one. This is because we believe, analogous to building a neural network in standard PyTorch, it is important to have the flexibility to include whatever components are necessary. As can be seen in more complicated examples, this allows the user great flexibility in designing custom models.
For most GP regression models, you will need to construct the following GPyTorch objects:
A GP Model (
gpytorch.models.ExactGP) - This handles most of the inference.A Likelihood (
gpytorch.likelihoods.GaussianLikelihood) - This is the most common likelihood used for GP regression.A Mean - This defines the prior mean of the GP.(If you don’t know which mean to use, a
gpytorch.means.ConstantMean()is a good place to start.)A Kernel - This defines the prior covariance of the GP.(If you don’t know which kernel to use, a
gpytorch.kernels.ScaleKernel(gpytorch.kernels.RBFKernel())is a good place to start).A MultivariateNormal Distribution (
gpytorch.distributions.MultivariateNormal) - This is the object used to represent multivariate normal distributions.
The GP Model¶
The components of a user built (Exact, i.e. non-variational) GP model in GPyTorch are, broadly speaking:
An
__init__method that takes the training data and a likelihood, and constructs whatever objects are necessary for the model’sforwardmethod. This will most commonly include things like a mean module and a kernel module.A
forwardmethod that takes in some \(n \times d\) dataxand returns aMultivariateNormalwith the prior mean and covariance evaluated atx. In other words, we return the vector \(\mu(x)\) and the \(n \times n\) matrix \(K_{xx}\) representing the prior mean and covariance matrix of the GP.
This specification leaves a large amount of flexibility when defining a model. For example, to compose two kernels via addition, you can either add the kernel modules directly:
self.covar_module = ScaleKernel(RBFKernel() + LinearKernel())
Or you can add the outputs of the kernel in the forward method:
covar_x = self.rbf_kernel_module(x) + self.white_noise_module(x)
The likelihood¶
The simplest likelihood for regression is the gpytorch.likelihoods.GaussianLikelihood. This assumes a homoskedastic noise model (i.e. all inputs have the same observational noise).
There are other options for exact GP regression, such as the FixedNoiseGaussianLikelihood, which assigns a different observed noise value to different training inputs.
[9]:
# We will use the simplest form of GP model, exact inference
class ExactGPModel(gpytorch.models.ExactGP):
def __init__(self, train_x, train_y, likelihood):
super(ExactGPModel, self).__init__(train_x, train_y, likelihood)
self.mean_module = gpytorch.means.ConstantMean()
self.covar_module = gpytorch.kernels.ScaleKernel(gpytorch.kernels.RBFKernel())
def forward(self, x):
mean_x = self.mean_module(x)
covar_x = self.covar_module(x)
return gpytorch.distributions.MultivariateNormal(mean_x, covar_x)
# initialize likelihood and model
likelihood = gpytorch.likelihoods.GaussianLikelihood()
model = ExactGPModel(train_x, train_y, likelihood)
Model modes¶
Like most PyTorch modules, the ExactGP has a .train() and .eval() mode. - .train() mode is for optimizing model hyperameters. - .eval() mode is for computing predictions through the model posterior.
Training the model¶
In the next cell, we handle using Type-II MLE to train the hyperparameters of the Gaussian process.
The most obvious difference here compared to many other GP implementations is that, as in standard PyTorch, the core training loop is written by the user. In GPyTorch, we make use of the standard PyTorch optimizers as from torch.optim, and all trainable parameters of the model should be of type torch.nn.Parameter. Because GP models directly extend torch.nn.Module, calls to methods like model.parameters() or model.named_parameters() function as you might expect coming from
PyTorch.
In most cases, the boilerplate code below will work well. It has the same basic components as the standard PyTorch training loop:
Zero all parameter gradients
Call the model and compute the loss
Call backward on the loss to fill in gradients
Take a step on the optimizer
However, defining custom training loops allows for greater flexibility. For example, it is easy to save the parameters at each step of training, or use different learning rates for different parameters (which may be useful in deep kernel learning for example).
[10]:
# this is for running the notebook in our testing framework
import os
smoke_test = ('CI' in os.environ)
training_iter = 2 if smoke_test else 50
# Find optimal model hyperparameters
model.train()
likelihood.train()
# Use the adam optimizer
optimizer = torch.optim.Adam(model.parameters(), lr=0.1) # Includes GaussianLikelihood parameters
# "Loss" for GPs - the marginal log likelihood
mll = gpytorch.mlls.ExactMarginalLogLikelihood(likelihood, model)
for i in range(training_iter):
# Zero gradients from previous iteration
optimizer.zero_grad()
# Output from model
output = model(train_x)
# Calc loss and backprop gradients
loss = -mll(output, train_y)
loss.backward()
print('Iter %d/%d - Loss: %.3f lengthscale: %.3f noise: %.3f' % (
i + 1, training_iter, loss.item(),
model.covar_module.base_kernel.lengthscale.item(),
model.likelihood.noise.item()
))
optimizer.step()
Iter 1/50 - Loss: 0.939 lengthscale: 0.693 noise: 0.693
Iter 2/50 - Loss: 0.908 lengthscale: 0.644 noise: 0.644
Iter 3/50 - Loss: 0.874 lengthscale: 0.598 noise: 0.598
Iter 4/50 - Loss: 0.837 lengthscale: 0.555 noise: 0.554
Iter 5/50 - Loss: 0.795 lengthscale: 0.514 noise: 0.513
Iter 6/50 - Loss: 0.749 lengthscale: 0.476 noise: 0.474
Iter 7/50 - Loss: 0.699 lengthscale: 0.440 noise: 0.437
Iter 8/50 - Loss: 0.649 lengthscale: 0.405 noise: 0.402
Iter 9/50 - Loss: 0.600 lengthscale: 0.372 noise: 0.369
Iter 10/50 - Loss: 0.556 lengthscale: 0.342 noise: 0.339
Iter 11/50 - Loss: 0.516 lengthscale: 0.315 noise: 0.310
Iter 12/50 - Loss: 0.480 lengthscale: 0.291 noise: 0.284
Iter 13/50 - Loss: 0.448 lengthscale: 0.270 noise: 0.259
Iter 14/50 - Loss: 0.413 lengthscale: 0.254 noise: 0.237
Iter 15/50 - Loss: 0.380 lengthscale: 0.241 noise: 0.216
Iter 16/50 - Loss: 0.355 lengthscale: 0.231 noise: 0.197
Iter 17/50 - Loss: 0.314 lengthscale: 0.223 noise: 0.179
Iter 18/50 - Loss: 0.292 lengthscale: 0.218 noise: 0.163
Iter 19/50 - Loss: 0.262 lengthscale: 0.214 noise: 0.148
Iter 20/50 - Loss: 0.236 lengthscale: 0.214 noise: 0.135
Iter 21/50 - Loss: 0.201 lengthscale: 0.216 noise: 0.122
Iter 22/50 - Loss: 0.176 lengthscale: 0.220 noise: 0.111
Iter 23/50 - Loss: 0.158 lengthscale: 0.224 noise: 0.102
Iter 24/50 - Loss: 0.125 lengthscale: 0.231 noise: 0.093
Iter 25/50 - Loss: 0.101 lengthscale: 0.239 noise: 0.085
Iter 26/50 - Loss: 0.078 lengthscale: 0.247 noise: 0.077
Iter 27/50 - Loss: 0.066 lengthscale: 0.256 noise: 0.071
Iter 28/50 - Loss: 0.052 lengthscale: 0.265 noise: 0.065
Iter 29/50 - Loss: 0.036 lengthscale: 0.276 noise: 0.060
Iter 30/50 - Loss: 0.036 lengthscale: 0.286 noise: 0.056
Iter 31/50 - Loss: 0.031 lengthscale: 0.297 noise: 0.052
Iter 32/50 - Loss: 0.028 lengthscale: 0.306 noise: 0.048
Iter 33/50 - Loss: 0.030 lengthscale: 0.315 noise: 0.045
Iter 34/50 - Loss: 0.035 lengthscale: 0.322 noise: 0.043
Iter 35/50 - Loss: 0.039 lengthscale: 0.326 noise: 0.041
Iter 36/50 - Loss: 0.043 lengthscale: 0.329 noise: 0.039
Iter 37/50 - Loss: 0.047 lengthscale: 0.327 noise: 0.038
Iter 38/50 - Loss: 0.052 lengthscale: 0.323 noise: 0.037
Iter 39/50 - Loss: 0.048 lengthscale: 0.317 noise: 0.036
Iter 40/50 - Loss: 0.051 lengthscale: 0.309 noise: 0.036
Iter 41/50 - Loss: 0.051 lengthscale: 0.302 noise: 0.036
Iter 42/50 - Loss: 0.047 lengthscale: 0.295 noise: 0.036
Iter 43/50 - Loss: 0.048 lengthscale: 0.288 noise: 0.036
Iter 44/50 - Loss: 0.047 lengthscale: 0.281 noise: 0.037
Iter 45/50 - Loss: 0.047 lengthscale: 0.276 noise: 0.037
Iter 46/50 - Loss: 0.040 lengthscale: 0.273 noise: 0.038
Iter 47/50 - Loss: 0.037 lengthscale: 0.271 noise: 0.039
Iter 48/50 - Loss: 0.040 lengthscale: 0.270 noise: 0.040
Iter 49/50 - Loss: 0.033 lengthscale: 0.269 noise: 0.042
Iter 50/50 - Loss: 0.032 lengthscale: 0.269 noise: 0.043
Make predictions with the model¶
In the next cell, we make predictions with the model. To do this, we simply put the model and likelihood in eval mode, and call both modules on the test data.
Just as a user defined GP model returns a MultivariateNormal containing the prior mean and covariance from forward, a trained GP model in eval mode returns a MultivariateNormal containing the posterior mean and covariance.
If we denote a test point (test_x) as x* with the true output being y*, then model(test_x) returns the model posterior distribution p(f* | x*, X, y), for training data X, y. This posterior is the distribution over the function we are trying to model, and thus quantifies our model uncertainty.
In contrast, likelihood(model(test_x)) gives us the posterior predictive distribution p(y* | x*, X, y) which is the probability distribution over the predicted output value. Recall in our problem setup
where 𝜖 is the likelihood noise for each observation. By including the likelihood noise which is the noise in your observation (e.g. due to noisy sensor), the prediction is over the observed value of the test point.
Thus, getting the predictive mean and variance, and then sampling functions from the GP at the given test points could be accomplished with calls like:
f_preds = model(test_x)
y_preds = likelihood(model(test_x))
f_mean = f_preds.mean
f_var = f_preds.variance
f_covar = f_preds.covariance_matrix
f_samples = f_preds.sample(sample_shape=torch.Size(1000,))
The gpytorch.settings.fast_pred_var context is not needed, but here we are giving a preview of using one of our cool features, getting faster predictive distributions using LOVE.
[11]:
# Get into evaluation (predictive posterior) mode
model.eval()
likelihood.eval()
# Test points are regularly spaced along [0,1]
# Make predictions by feeding model through likelihood
with torch.no_grad(), gpytorch.settings.fast_pred_var():
test_x = torch.linspace(0, 1, 51)
observed_pred = likelihood(model(test_x))
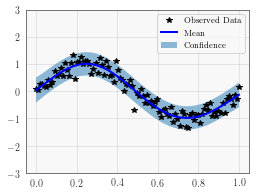
Plot the model fit¶
In the next cell, we plot the mean and confidence region of the Gaussian process model. The confidence_region method is a helper method that returns 2 standard deviations above and below the mean.
[12]:
with torch.no_grad():
# Initialize plot
f, ax = plt.subplots(1, 1, figsize=(4, 3))
# Get upper and lower confidence bounds
lower, upper = observed_pred.confidence_region()
# Plot training data as black stars
ax.plot(train_x.numpy(), train_y.numpy(), 'k*')
# Plot predictive means as blue line
ax.plot(test_x.numpy(), observed_pred.mean.numpy(), 'b')
# Shade between the lower and upper confidence bounds
ax.fill_between(test_x.numpy(), lower.numpy(), upper.numpy(), alpha=0.5)
ax.set_ylim([-3, 3])
ax.legend(['Observed Data', 'Mean', 'Confidence'])