GPyTorch Regression Tutorial (GPU)¶
(This notebook is the same as the simple GP regression tutorial notebook, but does all computations on a GPU for acceleration. Check out the multi-GPU tutorial if you have large datasets that needs multiple GPUs!)
Introduction¶
In this notebook, we demonstrate many of the design features of GPyTorch using the simplest example, training an RBF kernel Gaussian process on a simple function. We’ll be modeling the function
with 11 training examples, and testing on 51 test examples.
[1]:
import math
import torch
import gpytorch
from matplotlib import pyplot as plt
%matplotlib inline
%load_ext autoreload
%autoreload 2
Set up training data¶
In the next cell, we set up the training data for this example. We’ll be using 11 regularly spaced points on [0,1] which we evaluate the function on and add Gaussian noise to get the training labels.
[2]:
# Training data is 11 points in [0,1] inclusive regularly spaced
train_x = torch.linspace(0, 1, 100)
# True function is sin(2*pi*x) with Gaussian noise
train_y = torch.sin(train_x * (2 * math.pi)) + torch.randn(train_x.size()) * 0.2
Setting up the model¶
See the simple GP regression tutorial for a detailed explanation for all the terms.
[3]:
class ExactGPModel(gpytorch.models.ExactGP):
def __init__(self, train_x, train_y, likelihood):
super(ExactGPModel, self).__init__(train_x, train_y, likelihood)
self.mean_module = gpytorch.means.ConstantMean()
self.covar_module = gpytorch.kernels.ScaleKernel(gpytorch.kernels.RBFKernel())
def forward(self, x):
mean_x = self.mean_module(x)
covar_x = self.covar_module(x)
return gpytorch.distributions.MultivariateNormal(mean_x, covar_x)
# initialize likelihood and model
likelihood = gpytorch.likelihoods.GaussianLikelihood()
model = ExactGPModel(train_x, train_y, likelihood)
Using the GPU¶
To do computations on the GPU, we need to put our data and model onto the GPU. (This requires PyTorch with CUDA).
[4]:
train_x = train_x.cuda()
train_y = train_y.cuda()
model = model.cuda()
likelihood = likelihood.cuda()
That’s it! All the training code is the same as in the simple GP regression tutorial.
Training the model¶
[5]:
# Find optimal model hyperparameters
model.train()
likelihood.train()
# Use the adam optimizer
optimizer = torch.optim.Adam(model.parameters(), lr=0.1) # Includes GaussianLikelihood parameters
# "Loss" for GPs - the marginal log likelihood
mll = gpytorch.mlls.ExactMarginalLogLikelihood(likelihood, model)
training_iter = 50
for i in range(training_iter):
# Zero gradients from previous iteration
optimizer.zero_grad()
# Output from model
output = model(train_x)
# Calc loss and backprop gradients
loss = -mll(output, train_y)
loss.backward()
print('Iter %d/%d - Loss: %.3f lengthscale: %.3f noise: %.3f' % (
i + 1, training_iter, loss.item(),
model.covar_module.base_kernel.lengthscale.item(),
model.likelihood.noise.item()
))
optimizer.step()
Iter 1/50 - Loss: 0.944 lengthscale: 0.693 noise: 0.693
Iter 2/50 - Loss: 0.913 lengthscale: 0.644 noise: 0.644
Iter 3/50 - Loss: 0.879 lengthscale: 0.598 noise: 0.598
Iter 4/50 - Loss: 0.841 lengthscale: 0.555 noise: 0.554
Iter 5/50 - Loss: 0.798 lengthscale: 0.514 noise: 0.513
Iter 6/50 - Loss: 0.750 lengthscale: 0.475 noise: 0.474
Iter 7/50 - Loss: 0.698 lengthscale: 0.439 noise: 0.437
Iter 8/50 - Loss: 0.645 lengthscale: 0.405 noise: 0.402
Iter 9/50 - Loss: 0.595 lengthscale: 0.372 noise: 0.369
Iter 10/50 - Loss: 0.548 lengthscale: 0.342 noise: 0.339
Iter 11/50 - Loss: 0.507 lengthscale: 0.315 noise: 0.310
Iter 12/50 - Loss: 0.469 lengthscale: 0.292 noise: 0.284
Iter 13/50 - Loss: 0.432 lengthscale: 0.272 noise: 0.259
Iter 14/50 - Loss: 0.398 lengthscale: 0.255 noise: 0.236
Iter 15/50 - Loss: 0.363 lengthscale: 0.241 noise: 0.215
Iter 16/50 - Loss: 0.329 lengthscale: 0.230 noise: 0.196
Iter 17/50 - Loss: 0.296 lengthscale: 0.222 noise: 0.178
Iter 18/50 - Loss: 0.263 lengthscale: 0.215 noise: 0.162
Iter 19/50 - Loss: 0.230 lengthscale: 0.210 noise: 0.147
Iter 20/50 - Loss: 0.198 lengthscale: 0.207 noise: 0.134
Iter 21/50 - Loss: 0.167 lengthscale: 0.205 noise: 0.122
Iter 22/50 - Loss: 0.136 lengthscale: 0.205 noise: 0.110
Iter 23/50 - Loss: 0.107 lengthscale: 0.206 noise: 0.100
Iter 24/50 - Loss: 0.079 lengthscale: 0.208 noise: 0.091
Iter 25/50 - Loss: 0.053 lengthscale: 0.211 noise: 0.083
Iter 26/50 - Loss: 0.028 lengthscale: 0.215 noise: 0.076
Iter 27/50 - Loss: 0.006 lengthscale: 0.220 noise: 0.069
Iter 28/50 - Loss: -0.013 lengthscale: 0.225 noise: 0.063
Iter 29/50 - Loss: -0.029 lengthscale: 0.231 noise: 0.058
Iter 30/50 - Loss: -0.043 lengthscale: 0.237 noise: 0.053
Iter 31/50 - Loss: -0.053 lengthscale: 0.243 noise: 0.049
Iter 32/50 - Loss: -0.060 lengthscale: 0.249 noise: 0.045
Iter 33/50 - Loss: -0.065 lengthscale: 0.254 noise: 0.042
Iter 34/50 - Loss: -0.066 lengthscale: 0.259 noise: 0.039
Iter 35/50 - Loss: -0.066 lengthscale: 0.262 noise: 0.037
Iter 36/50 - Loss: -0.063 lengthscale: 0.265 noise: 0.035
Iter 37/50 - Loss: -0.060 lengthscale: 0.266 noise: 0.033
Iter 38/50 - Loss: -0.056 lengthscale: 0.266 noise: 0.032
Iter 39/50 - Loss: -0.052 lengthscale: 0.265 noise: 0.031
Iter 40/50 - Loss: -0.049 lengthscale: 0.262 noise: 0.030
Iter 41/50 - Loss: -0.047 lengthscale: 0.259 noise: 0.029
Iter 42/50 - Loss: -0.046 lengthscale: 0.254 noise: 0.029
Iter 43/50 - Loss: -0.046 lengthscale: 0.249 noise: 0.029
Iter 44/50 - Loss: -0.047 lengthscale: 0.243 noise: 0.029
Iter 45/50 - Loss: -0.049 lengthscale: 0.237 noise: 0.029
Iter 46/50 - Loss: -0.051 lengthscale: 0.231 noise: 0.029
Iter 47/50 - Loss: -0.054 lengthscale: 0.225 noise: 0.030
Iter 48/50 - Loss: -0.057 lengthscale: 0.219 noise: 0.030
Iter 49/50 - Loss: -0.059 lengthscale: 0.214 noise: 0.031
Iter 50/50 - Loss: -0.061 lengthscale: 0.210 noise: 0.032
Make predictions with the model¶
First, we need to make some test data, and then throw it onto the GPU
[6]:
test_x = torch.linspace(0, 1, 51).cuda()
Now the rest of the code follows the simple GP regression tutorial.
[7]:
# Get into evaluation (predictive posterior) mode
model.eval()
likelihood.eval()
# Test points are regularly spaced along [0,1]
# Make predictions by feeding model through likelihood
with torch.no_grad(), gpytorch.settings.fast_pred_var():
observed_pred = likelihood(model(test_x))
mean = observed_pred.mean
lower, upper = observed_pred.confidence_region()
For plotting, we’re going to grab the data from the GPU and put it back on the CPU. We can accomplish this with the .cpu() function.
[8]:
mean = mean.cpu()
lower = lower.cpu()
upper = upper.cpu()
train_x = train_x.cpu()
train_y = train_y.cpu()
test_x = test_x.cpu()
[9]:
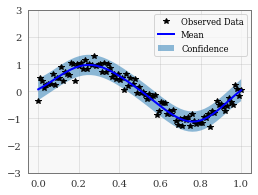
with torch.no_grad():
# Initialize plot
f, ax = plt.subplots(1, 1, figsize=(4, 3))
# Plot training data as black stars
ax.plot(train_x.numpy(), train_y.numpy(), 'k*')
# Plot predictive means as blue line
ax.plot(test_x.numpy(), mean.numpy(), 'b')
# Shade between the lower and upper confidence bounds
ax.fill_between(test_x.numpy(), lower.numpy(), upper.numpy(), alpha=0.5)
ax.set_ylim([-3, 3])
ax.legend(['Observed Data', 'Mean', 'Confidence'])