Metrics in GPyTorch¶
In this notebook, we will see how to evaluate GPyTorch models with probabilistic metrics.
Note: It is encouraged to check the Simple GP Regression notebook first if not done already. We’ll reuse most of the code from there.
We’ll be modeling the function
[26]:
import math
import torch
import gpytorch
from matplotlib import pyplot as plt
%matplotlib inline
%load_ext autoreload
%autoreload 2
The autoreload extension is already loaded. To reload it, use:
%reload_ext autoreload
In the next cell, we set up the train and test data.
[27]:
# Training data is 100 points in [0,1] inclusive regularly spaced
train_x = torch.linspace(0, 1, 100)
# True function is sin(2*pi*x) with Gaussian noise
train_y = torch.sin(train_x * (2 * math.pi)) + torch.randn(train_x.size()) * math.sqrt(0.04)
test_x = torch.linspace(0, 1, 51)
test_y = torch.sin(test_x * (2 * math.pi)) + torch.randn(test_x.size()) * math.sqrt(0.04)
In the next cell, we define a simple GP regression model.
[28]:
# We will use the simplest form of GP model, exact inference
class ExactGPModel(gpytorch.models.ExactGP):
def __init__(self, train_x, train_y, likelihood):
super(ExactGPModel, self).__init__(train_x, train_y, likelihood)
self.mean_module = gpytorch.means.ConstantMean()
self.covar_module = gpytorch.kernels.ScaleKernel(gpytorch.kernels.RBFKernel())
def forward(self, x):
mean_x = self.mean_module(x)
covar_x = self.covar_module(x)
return gpytorch.distributions.MultivariateNormal(mean_x, covar_x)
# initialize likelihood and model
likelihood = gpytorch.likelihoods.GaussianLikelihood()
model = ExactGPModel(train_x, train_y, likelihood)
Our model is ready for hyperparameter learning, but, first let us check how it performs on the test data.
[29]:
model.eval()
with torch.no_grad():
untrained_pred_dist = likelihood(model(test_x))
predictive_mean = untrained_pred_dist.mean
lower, upper = untrained_pred_dist.confidence_region()
f, ax = plt.subplots(1, 1, figsize=(4, 3))
# Plot training data as black stars
ax.plot(train_x, train_y, 'k*')
# Plot predictive means as blue line
ax.plot(test_x, predictive_mean, 'b')
# Shade between the lower and upper confidence bounds
ax.fill_between(test_x, lower, upper, alpha=0.5)
ax.set_ylim([-3, 3])
ax.legend(['Observed Data', 'Mean', 'Confidence'], bbox_to_anchor=(1.6,1));
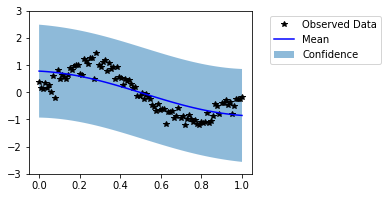
Visually, this does not look like a good fit. Now, let us train the model hyperparameters.
[30]:
# this is for running the notebook in our testing framework
import os
smoke_test = ('CI' in os.environ)
training_iter = 2 if smoke_test else 50
# Find optimal model hyperparameters
model.train()
# Use the adam optimizer
optimizer = torch.optim.Adam(model.parameters(), lr=0.1) # Includes GaussianLikelihood parameters
# "Loss" for GPs - the marginal log likelihood
mll = gpytorch.mlls.ExactMarginalLogLikelihood(likelihood, model)
for i in range(training_iter):
# Zero gradients from previous iteration
optimizer.zero_grad()
# Output from model
output = model(train_x)
# Calc loss and backprop gradients
loss = -mll(output, train_y)
loss.backward()
print('Iter %d/%d - Loss: %.3f lengthscale: %.3f noise: %.3f' % (
i + 1, training_iter, loss.item(),
model.covar_module.base_kernel.lengthscale.item(),
model.likelihood.noise.item()
))
optimizer.step()
Iter 1/50 - Loss: 0.944 lengthscale: 0.693 noise: 0.693
Iter 2/50 - Loss: 0.913 lengthscale: 0.644 noise: 0.644
Iter 3/50 - Loss: 0.879 lengthscale: 0.598 noise: 0.598
Iter 4/50 - Loss: 0.841 lengthscale: 0.555 noise: 0.554
Iter 5/50 - Loss: 0.798 lengthscale: 0.514 noise: 0.513
Iter 6/50 - Loss: 0.750 lengthscale: 0.475 noise: 0.474
Iter 7/50 - Loss: 0.698 lengthscale: 0.439 noise: 0.437
Iter 8/50 - Loss: 0.645 lengthscale: 0.404 noise: 0.402
Iter 9/50 - Loss: 0.594 lengthscale: 0.372 noise: 0.369
Iter 10/50 - Loss: 0.547 lengthscale: 0.342 noise: 0.339
Iter 11/50 - Loss: 0.505 lengthscale: 0.315 noise: 0.310
Iter 12/50 - Loss: 0.466 lengthscale: 0.291 noise: 0.284
Iter 13/50 - Loss: 0.430 lengthscale: 0.271 noise: 0.259
Iter 14/50 - Loss: 0.395 lengthscale: 0.254 noise: 0.236
Iter 15/50 - Loss: 0.361 lengthscale: 0.240 noise: 0.215
Iter 16/50 - Loss: 0.327 lengthscale: 0.229 noise: 0.196
Iter 17/50 - Loss: 0.294 lengthscale: 0.220 noise: 0.178
Iter 18/50 - Loss: 0.261 lengthscale: 0.214 noise: 0.162
Iter 19/50 - Loss: 0.228 lengthscale: 0.209 noise: 0.147
Iter 20/50 - Loss: 0.196 lengthscale: 0.206 noise: 0.134
Iter 21/50 - Loss: 0.164 lengthscale: 0.205 noise: 0.122
Iter 22/50 - Loss: 0.133 lengthscale: 0.204 noise: 0.110
Iter 23/50 - Loss: 0.104 lengthscale: 0.205 noise: 0.100
Iter 24/50 - Loss: 0.076 lengthscale: 0.207 noise: 0.091
Iter 25/50 - Loss: 0.049 lengthscale: 0.210 noise: 0.083
Iter 26/50 - Loss: 0.025 lengthscale: 0.214 noise: 0.076
Iter 27/50 - Loss: 0.003 lengthscale: 0.218 noise: 0.069
Iter 28/50 - Loss: -0.017 lengthscale: 0.223 noise: 0.063
Iter 29/50 - Loss: -0.033 lengthscale: 0.228 noise: 0.058
Iter 30/50 - Loss: -0.047 lengthscale: 0.233 noise: 0.053
Iter 31/50 - Loss: -0.057 lengthscale: 0.239 noise: 0.049
Iter 32/50 - Loss: -0.065 lengthscale: 0.243 noise: 0.045
Iter 33/50 - Loss: -0.069 lengthscale: 0.248 noise: 0.042
Iter 34/50 - Loss: -0.071 lengthscale: 0.252 noise: 0.039
Iter 35/50 - Loss: -0.071 lengthscale: 0.255 noise: 0.037
Iter 36/50 - Loss: -0.069 lengthscale: 0.257 noise: 0.035
Iter 37/50 - Loss: -0.066 lengthscale: 0.257 noise: 0.033
Iter 38/50 - Loss: -0.062 lengthscale: 0.257 noise: 0.032
Iter 39/50 - Loss: -0.058 lengthscale: 0.255 noise: 0.031
Iter 40/50 - Loss: -0.055 lengthscale: 0.252 noise: 0.030
Iter 41/50 - Loss: -0.054 lengthscale: 0.248 noise: 0.029
Iter 42/50 - Loss: -0.053 lengthscale: 0.243 noise: 0.029
Iter 43/50 - Loss: -0.053 lengthscale: 0.237 noise: 0.029
Iter 44/50 - Loss: -0.055 lengthscale: 0.230 noise: 0.029
Iter 45/50 - Loss: -0.057 lengthscale: 0.223 noise: 0.029
Iter 46/50 - Loss: -0.060 lengthscale: 0.216 noise: 0.029
Iter 47/50 - Loss: -0.063 lengthscale: 0.208 noise: 0.030
Iter 48/50 - Loss: -0.066 lengthscale: 0.202 noise: 0.030
Iter 49/50 - Loss: -0.068 lengthscale: 0.196 noise: 0.031
Iter 50/50 - Loss: -0.070 lengthscale: 0.190 noise: 0.032
In the next cell, we reevaluate the model on the test data.
[31]:
model.eval()
with torch.no_grad():
trained_pred_dist = likelihood(model(test_x))
predictive_mean = trained_pred_dist.mean
lower, upper = trained_pred_dist.confidence_region()
f, ax = plt.subplots(1, 1, figsize=(4, 3))
# Plot training data as black stars
ax.plot(train_x, train_y, 'k*')
# Plot predictive means as blue line
ax.plot(test_x, predictive_mean, 'b')
# Shade between the lower and upper confidence bounds
ax.fill_between(test_x, lower, upper, alpha=0.5)
ax.set_ylim([-3, 3])
ax.legend(['Observed Data', 'Mean', 'Confidence'], bbox_to_anchor=(1.6,1));
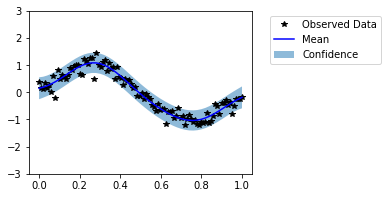
Now our model seems to fit well on the data. It is not always possible to visually evaluate the model in high dimensional cases. Thus, now we evaluate the models with help of probabilistic metrics. We have saved predictive distributions from untrained and trained models as untrained_pred_dist and trained_pred_dist respectively.
Negative Log Predictive Density (NLPD)¶
Negative Log Predictive Density (NLPD) is the most standard probabilistic metric for evaluating GP models. In simple terms, it is negative log likelihood of the test data given the predictive distribution. It can be computed as follows:
[32]:
init_nlpd = gpytorch.metrics.negative_log_predictive_density(untrained_pred_dist, test_y)
final_nlpd = gpytorch.metrics.negative_log_predictive_density(trained_pred_dist, test_y)
print(f'Untrained model NLPD: {init_nlpd:.2f}, \nTrained model NLPD: {final_nlpd:.2f}')
Untrained model NLPD: 0.88,
Trained model NLPD: -0.21
Mean Standardized Log Loss (MSLL)¶
This metric computes average negative log likelihood of all test points w.r.t their univariate predicitve densities. For more details, see “page No. 23, Gaussian Processes for Machine Learning, Carl Edward Rasmussen and Christopher K. I. Williams, The MIT Press, 2006. ISBN 0-262-18253-X”
[33]:
init_msll = gpytorch.metrics.mean_standardized_log_loss(untrained_pred_dist, test_y)
final_msll = gpytorch.metrics.mean_standardized_log_loss(trained_pred_dist, test_y)
print(f'Untrained model MSLL: {init_msll:.2f}, \nTrained model MSLL: {final_msll:.2f}')
Untrained model MSLL: 0.82,
Trained model MSLL: -0.49
It is also possible to calculate the quantile coverage error with gpytorch.metrics.quantile_coverage_error function.
[35]:
quantile = 95
qce = gpytorch.metrics.quantile_coverage_error(trained_pred_dist, test_y, quantile=quantile)
print(f'Quantile {quantile}% Coverage Error: {qce:.2f}')
Quantile 95% Coverage Error: 0.01
Mean Squared Error (MSE)¶
Mean Squared Error (MSE) is the mean of the squared difference between the test observations and the predictive mean. It is a well-known conventional metric for evaluating regression models. However, it can not take uncertainty into account unlike NLPD, MLSS and ACE.
[36]:
init_mse = gpytorch.metrics.mean_squared_error(untrained_pred_dist, test_y, squared=True)
final_mse = gpytorch.metrics.mean_squared_error(trained_pred_dist, test_y, squared=True)
print(f'Untrained model MSE: {init_mse:.2f}, \nTrained model MSE: {final_mse:.2f}')
Untrained model MSE: 0.20,
Trained model MSE: 0.04
Mean Absolute Error (MAE)¶
Mean Absolute Error (MAE) is the mean of the absolute difference between the test observations and the predictive mean. It is less sensitive to the outliers than MSE.
[37]:
init_mae = gpytorch.metrics.mean_absolute_error(untrained_pred_dist, test_y)
final_mae = gpytorch.metrics.mean_absolute_error(trained_pred_dist, test_y)
print(f'Untrained model MAE: {init_mae:.2f}, \nTrained model MAE: {final_mae:.2f}')
Untrained model MAE: 0.38,
Trained model MSE: 0.16